【Structural and Functional Analysis of the Ribosomal Stalk Protein】
Translation is the process in which ribosomes link amino acids to synthesize
proteins, based on the genetic information copied in mRNA. Translation
is a complex reaction consisting of four stages: initiation, elongation,
termination, and recycling, and proceeds by the interactions of the ribosome
with individual translation factors that act in each stage. The ribosomal
stalk protein is a long, flexible structure that exists in multiple copies
on the large ribosomal subunit, and plays a crucial role in the interactions
between the ribosome and various translation factors (Fig. 1). To date, we have found that the ribosomal stalk protein directly
interacts with various GTPase and ATPase translators via its C-terminal
region. We have also shown that the ribosome stalk protein is essential
for recruiting translation factors to the ribosomal factor-binding center,
and for the activities of individual translation factors on the ribosome.

Fig. 1 Ribosomal stalk proteins.
However, the mechanisms by which the stalk protein interacts with various translation factors with different structures remain elusive. It is also unclear how the stalk protein recruits translation factors to the factor-binding center of the ribosome, and how it contributes to facilitating the actions of individual translation factors. To answer these open questions, we are performing X-ray crystallography and biochemical/molecular biological analyses (Fig. 2). We are conducting this project in collaboration with Dr. Toshio Uchiumi, Honorary Professor/Fellow of Niigata University (HP).
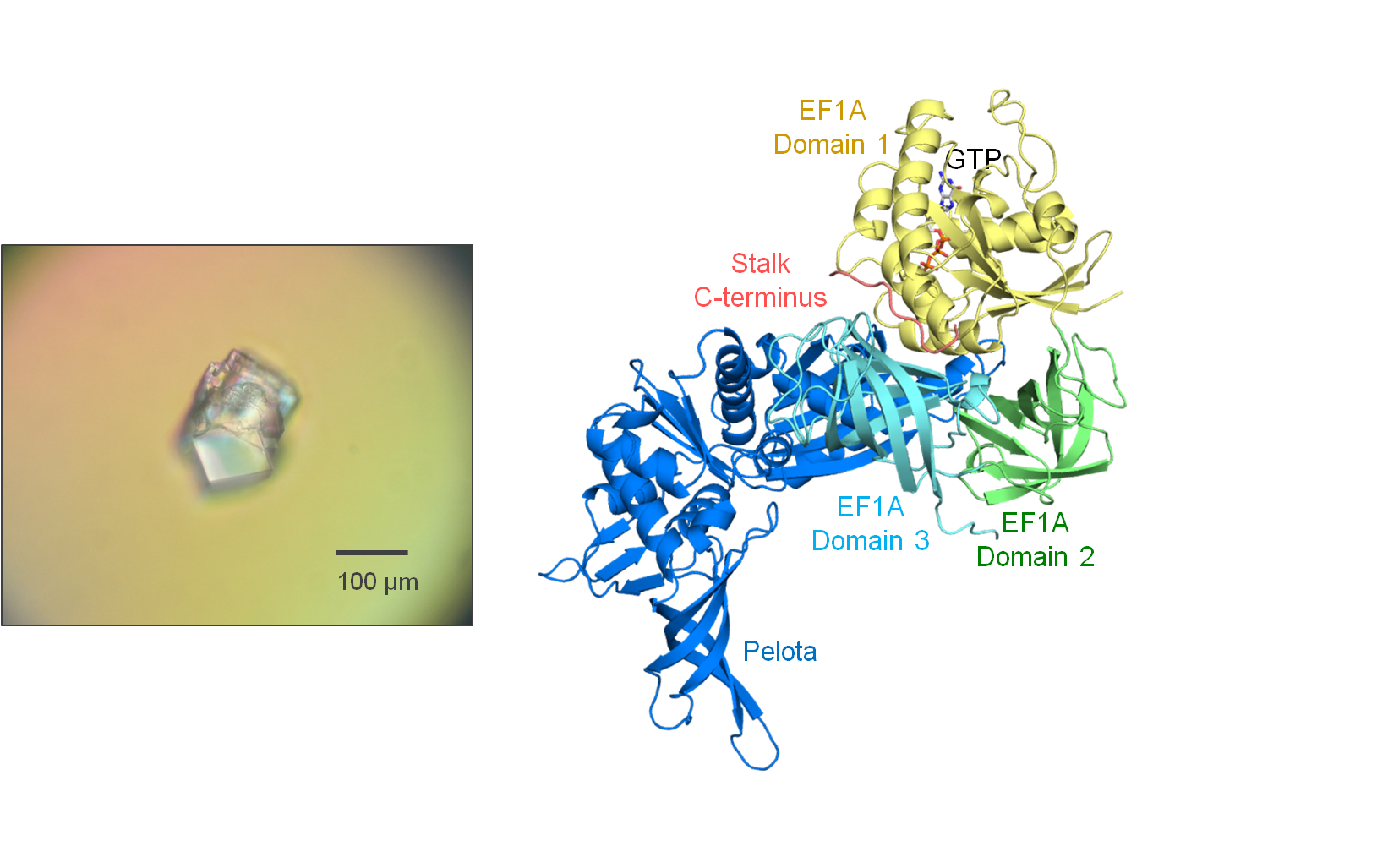
Fig. 2 Crystal (left) and structure (right) of the Ribosomal stalk protein•EF1A•Pelota
complex.
【Structural and Functional Analyses of Peptidyl-tRNA Hydrolase】
In translation, the peptide grows on the tRNA on the ribosome, and when
the ribosome reaches a stop codon, the elongated peptide is released from
the tRNA and translation normally terminates. However, due to amino acid
starvation, mRNA decay, and other phenomena, the ribosome may stall and
not reach the stop codon. In this case, stalled ribosomes release peptidyl-tRNAs,
tRNAs with the elongating peptide remaining attached to them, as the premature
products of protein synthesis. The accumulation of peptidyl-tRNAs is toxic
because it depletes the pool of tRNAs available for translation, thereby
halting protein synthesis. Peptidyl-tRNA hydrolase (Pth) is an enzyme that
cleaves the ester bond between the peptide and the tRNA of the peptidyl-tRNA
molecule, to recycle the tRNA for further rounds of protein synthesis (Fig. 3). Pth is present in all living organisms and is an essential protein in
bacteria.

Fig. 3 Action of peptidyl-tRNA hydrolase (Pth).
The amino acid and nucleotide sequences of peptidyl-tRNAs within cells
vary, and Pth recognizes these diverse peptidyl-tRNA species as substrates.
However, the mechanism for this sequence-independent recognition by Pth
remains obscure. The catalytic process of the hydrolysis reaction is also
unknown. Furthermore, it is unclear where Pth works in the cell. To answer
these questions, we are using X-ray crystallography, biochemical/molecular
biological analyses, and enzymatic analyses (Fig. 4). We are also performing molecular genetics research using yeast, in collaboration
with Dr. Shuh-ichi Nishikawa of the Faculty of Science/Graduate School
of Science and Technology, Niigata University (HP). Notably, Pth is in the spotlight as an attractive antibiotic target.
The results of this project, therefore, will provide important data for
the development research of new antibiotics.

Fig. 4 Crystal (left) and structure (right) of Pth•tRNA CCA-acceptor-TΨC domain complex.